Abstract
Source: ASBMB poster abstract and slides. See references below.
My contributions (high level)
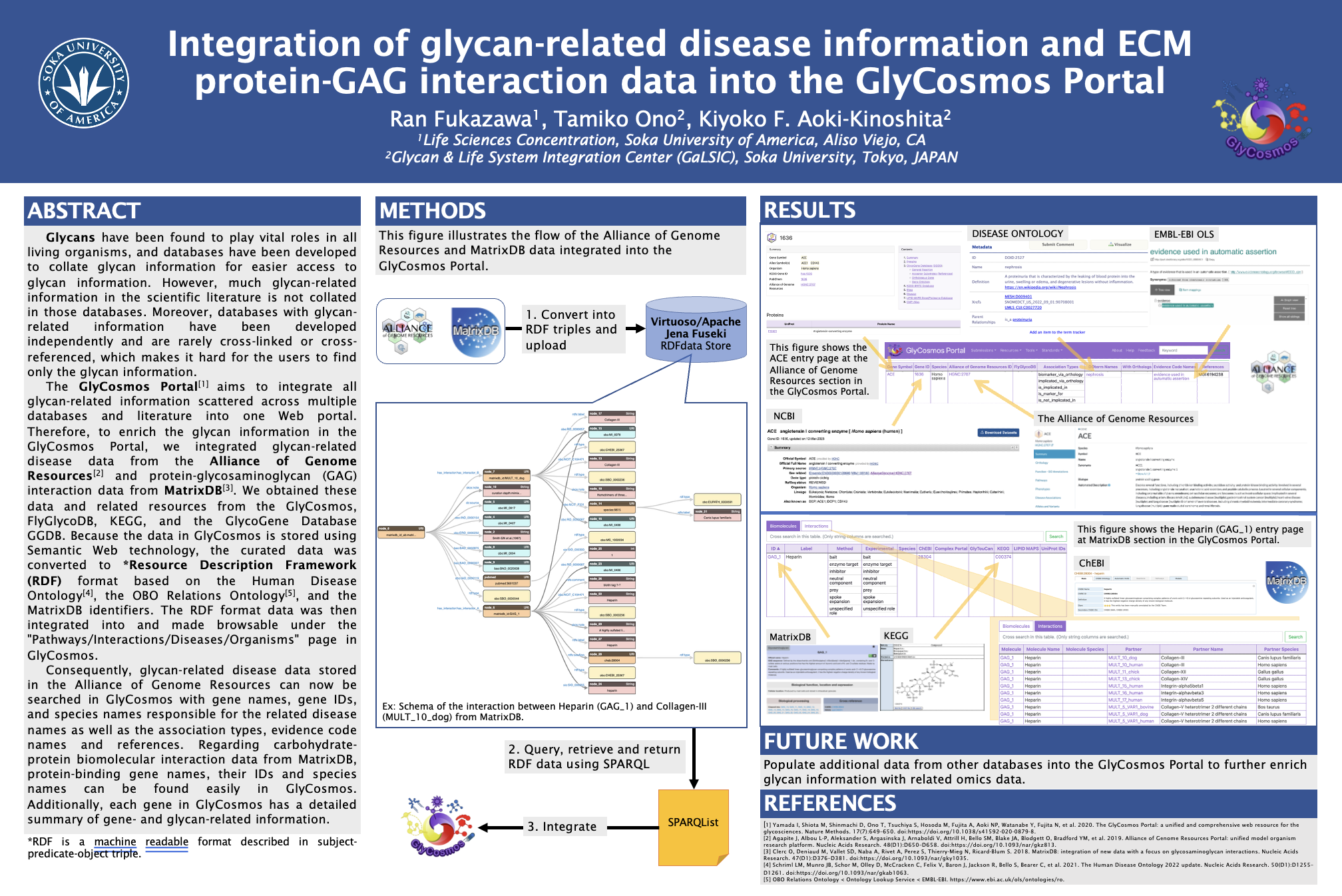
- Curated and mapped glycan-related disease records from the Alliance of Genome Resources into the GlyCosmos RDF data model.
- Converted MatrixDB protein–GAG interaction records into RDF triples using the Human Disease Ontology and MatrixDB identifiers.
- Integrated the converted data into GlyCosmos and added browse/search UI hooks under Pathways / Interactions / Diseases / Organisms.
- Authored SPARQL queries and helped verify results by cross-referencing KEGG, FlyGlycoDB and GGDB.
Technical notes
RDF store: Virtuoso / Apache Jena Fuseki
Query language: SPARQL
Ontologies: Disease Ontology, OBO RO
Sources: AGR, MatrixDB, KEGG, GGDB
How to reuse this page
Open this single HTML file in a browser. You can edit the abstract and add an embedded poster PDF or images in the #posterFrame area. The page includes a small JS utility to copy the abstract text and highlight key values.
Poster slide (preview)
References
- Yamada I, et al. The GlyCosmos Portal: a unified and comprehensive web resource for the glycosciences. Nature Methods (2020).
- Alliance of Genome Resources & MatrixDB sources (integrated for this work). See uploaded materials.